Getting started¶
In this tutorial we demonstrate how to create observables from raw data
Understanding the internal structure¶
pyobs is a library to manipulate observables. To do so a specific internal structure, transparent to the user, is defined and implemented to properly propagate errors among observables sharing data on the same configurations.
Observables defined from pyobs.observable contain 2 fundamental sub-classes
delta: contains the fluctuations on a given replica, ie on a set of configurations generated from a unique Markov Chain (stream). The fluctuations can be interpreted as master field if the appropriate arguments are passed.
cdata: contains the covariance matrix of observables for which the original fluctuations are not available, or that are obtained from different sources like experimental numbers.
When a new observable is created, one of the two sub-classes is automatically generated, while the others are left empty. During the analysis, when the different observables are combined together to extract new results, the library pyobs automatically takes care of combining data where it overlaps, ie taking correlations into account, and merging the sub-classes appropriately.
The sub-classes are collected in dictionaries, whose key uniquely defines either the pair (ensemble,replica) or covariance data. When different observables are combined together via some operation, these keys are used to identify the places where correlations have to be propagated.
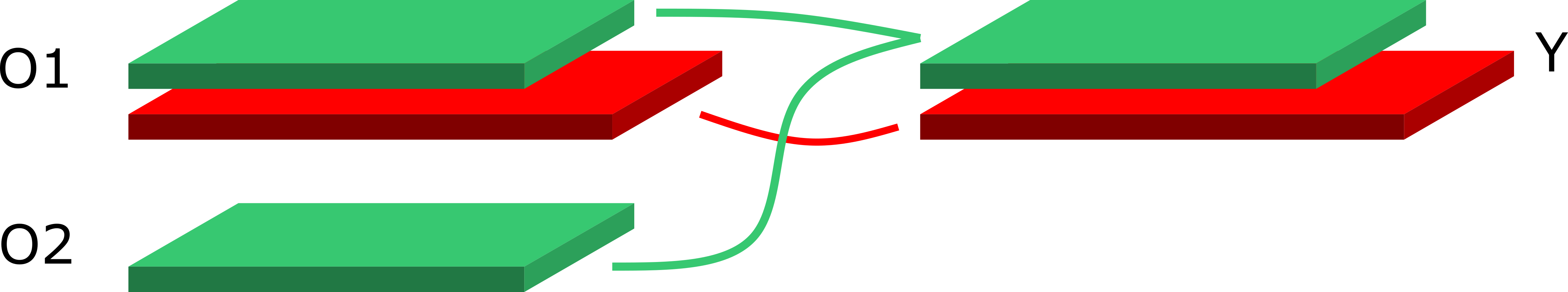
In the image above the green block represents a replica, with key green. When the two observables \(O_1, O_2\) are combined via a binary operation \(f(O_1, O_2) = Y\), correlations are properly propagated. The red block represents a replica with a different key (or a master-field or covariance data).
Creating the observable¶
In this example we generate synthetic autocorrelated data, using the pyobs.random module. In a generic situation we will have the measurement of a scalar quantity, for example, like the plaquette, on several gauge field configurations, stored in a python list, or in a numpy.array
[1]:
import pyobs
import numpy
import matplotlib.pyplot as plt
%matplotlib notebook
[2]:
N=5000 # number of configs
mu = 0.5 # central values
cov = (mu*0.5)**2 # error^2
# autocorrelation time
tau=4.0
rng = pyobs.random.generator('tutorial')
data = rng.markov_chain(mu,cov,tau,N)
Random generator initialized with seed = 649309182 [tutorial]
Then we create a new observable that we can inspect with the peek function.
[3]:
obsA = pyobs.observable(description='Observable A')
obsA.create('EnsA',data)
obsA.peek()
Observable with shape = (1,)
- description: Observable A
- created by mbruno at macthxbruno.local on Wed May 11 10:17:54 2022
- size: 218 KB
- mean: [0.02686853]
- Ensemble EnsA
- Replica 0 with ncnfg 5000
temporary additional memory required 0.076 MB
The first goal is to compute the error and check the autocorrelations. They should match \(\tau=4\).
[4]:
help(pyobs.observable.tauint)
obsA.tauint() # returns a dictionary with tauint ensemble by ensemble
Help on function tauint in module pyobs.core.ndobs:
tauint(self)
Estimates the integrated autocorrelation time and its error for every
ensemble, with the automatic windowing procedure.
Notes:
To be added in future versions: support for arbitrary values of Stau
[4]:
{'EnsA': [array([4.18879604]), array([0.60411676])]}
Below we print the error and plot it as a function of the summation window. The optimal value is chosen following U. Wolff’s algorithm.
[5]:
print(obsA) # this is equivalent to obsA.error()
[a,da] = obsA.error(plot=True) # note that a, da are numpy arrays
0.03(26)
A second observable¶
We generate a second observable that we imagine from a different ensemble.
[6]:
N=1000 # number of configs
mu = -2.5 # central values
cov = (mu*0.2)**2 # error^2
# autocorrelation time
tau=2.0
data = rng.markov_chain(mu,cov,tau,N)
obsB = pyobs.observable(description='Observable B')
obsB.create('EnsB',data)
obsB.error(plot=True);
When we perform an operation that combines the two observables, the fluctuations of both are properly propagated in the final result. When we use the plot=True argument, the contributions of the various ensembles to obsC are automatically plotted.
[7]:
obsC = obsA + 1./obsB
obsC.error(plot=True);
Multi-dimensional observables¶
As a final step, we examine how to create multi-dimensional observables, such as vectors, matrices and more complicated tensors. Let’s consider the case of a 2-D tensor. We should organize the data such that for every configuration the fastest index corresponds to columns
[8]:
# generate autocorrelated data
N=500 # number of configs
# central values
mu=[0.5, 1.2, -3.4, 60.2, -10.1, 5.01]
# diagonal cov matrix
cov=[(mu[i]*0.05)**2 for i in range(len(mu))]
# autocorrelation time
tau=4.0
data = rng.markov_chain(mu,cov,tau,N)
print(numpy.shape(data))
(500, 6)
[9]:
# create 2x3 observable, must flatten data
_data = numpy.zeros((N*2*3,))
for i in range(N):
for r in range(2):
for c in range(3):
_data[c+3*(r+2*i)] = data[i,c + 3*r] # fastest index column, slowest index configuration id
yobs = pyobs.observable()
yobs.create('ensA',_data,shape=(2,3))
print(yobs)
0.538(24) 1.147(72) -3.32(17)
56.3(3.0) -11.12(36) 5.10(25)
Creating master-field observables¶
In preparation
[ ]:
